Best practices for the safe transfer of rice germplasm
Contributers to this page: IRRI, Seed Health Unit, Los Banos, Philippines(Patria Gonzales, Evangeline Gonzales, Carlos Huelma, Myra Almodiel, Joel Dumlao).
The best practices in place at IRRI are summarized for key aspects related to the Seed Health Unit in this section.Preventing the introduction of diseases, weeds, and any potential pest to areas where these biological hazards are not present, is the primary function of any plant quarantine office of that location. General Guidelines for Importation, Use and Handling Wild Rice Seeds and/or Vegetative Parts are in place at IRRI.
Routine Seed Health Testing (RSHT) Quality and Quantity Standards include Blotter Test, Nematode Test, Macro Test/Tb cleaning, Documentation, Washing, Germination Test. Technical details on routine seed health testing procedures are provided.
|
ACTIVITY |
STANDARD QUANTITY (min qty/hr) |
STANDARD QUALITY |
|
1. Blotter Test a. Seeding
b. Evaluation
|
61-70 plates
Not Treated- 51-60 plates Treated -61-70 plates |
according to ISTA rules & standards (25 seeds/plate; readable/ correct labels; w/ 2 layers of moist blotter paper; seeds equidistant from one another & properly oriented
Accurate identification of seedborne organisms - no missed/misidentified organism in 99% plates evaluated; timely evaluation – 5 to 7 days after incubation. |
|
2. Nematode Test a. Setting/extraction
|
81-90 samples
51-65 samples |
correct weight of samples, correct amount of water (seeds are covered) germinated seeds are properly spread over the mesh wire, no seeds in the tygon tubing, pinchcock properly placed-no leakage, spillage of water/ seeds accurate identification (+/-; saprophytic or parasitic) and actual count (if parasitic) evaluation conducted as soon as extracts are gathered |
|
3. Macro Test/Tb cleaning |
1.1-1.5 K |
correct evaluation (+/-) of Tilletia barclayana |
|
4. Documentation |
181-200 samples |
data/info complete & accurate; manner (no erasures, readable) in which data is recorded generally good; immediately done after evaluation (consistency - 99% of the time) |
|
5. Washing |
241-260 plates; 76-85 funnels; 101-120 wire mesh
|
no pencil marks, water marks, fungal growth; thoroughly dried |
|
6. Germination Test
a. Seeding
b. Evaluation |
41-45 seedlots
|
correct number of seeds-100/tray; correct orientation of seeds, moisture of blotters, correct labelling;
timely with regards to evaluation (5 DAS, 7DAS, & 14 DAS); correct evaluation as to Normal/Abnormal seedlings, dead seeds (in accordance to ISTA Rules and Standards in Seedling Evaluation) |
At IRRI SHU, methods for the detection of fungal pathogens include: seed washing technique, blotter test, potato dextrose agar (PDA) method and the oatmeal agar method.
Methods for detection of fungal pathogens
1. Seed washing technique
This is useful in testing surface-borne, contaminating fungi like smuts, bunts, downy mildews, powdery mildews, rusts, etc.
Procedure
1. Place two grams of seed sample in a test tube and mix it well by adding 2 ml of sterile water for 5-10 minutes.
2. Centrifuge the supernatant solution at 200 rpm for 10 minutes and observe the sediments under a microscope for fungal structures.
2. Blotter test
Procedure
1. Line the lower lid of sterilized petri dishes with three layers of absorbent paper moistened with sterile water.
2. Drain off excess water and place 20–25 seeds manually with a forceps.
3. Evenly space the seeds to avoid contact with each other.
4. Incubate the seeds under near ultraviolet light in alternating cycles of 12-h light/darkness for 7 d at 20 + 2°C.
5. Examine the petri dishes under a stereo-binocular microscope for fungi developing on the seeds.
Profuse seedling growth may make interpretations difficult. This may be overcome by adding 2,4-D sodium salt to provide a 0.2% moistening solution.
3. Potato dextrose agar (PDA) method
Procedure
1. Prepare the medium by mixing 1 gram of Potato Dextrose Agar (PDA) powder in 100 ml of distilled water.
2. Sterilize the mixture in an autoclave for 15–20 min and cool to about 50°C.
3. Carefully pour the mixture into sterile petri dishes by lifting the lid enough only to pour in the agar to avoid contamination.
4. Allow it to cool and solidify for 20 min.
5. Surface-disinfect the seed by pretreating for 1 min in a 1% sodium hypochlorite (NaOCl) solution prepared by diluting 20 parts of laundry bleach (5.25% NaOCl) with 85 parts of water.
6. Place about 10 seeds (depending on size) on the agar surface with a forceps.
7. Incubate the petri dishes at 20–25°C for about 5–8 days.
8. Identify the seedborne pathogens on the basis of colony and spore characteristics.
Near ultraviolet light with a wavelength 300-380 nm (also called black light) may be required to promote sporulation.
Some times bacterial colonies develop on the agar and inhibit fungal growth making identification difficult. This can be overcome by adding an antibiotic such as streptomycin (500 ppm) to the autoclaved agar medium after it cools to 50–55°C.
4. Oatmeal agar method
Procedure
1. Prepare the medium by mixing 10 gram of ground oatmeal powder with 10g of pure agar in 1l of distilled water.
2. Warm on a stirrer hotplate mixing continuously until the agar has melted.
3. Sterilize the mixture in an autoclave at 220°C for 20 min and cool to about 50°C.
4. Mix in 4ml of tetracycline antibiotic solution (0.25g in 100ml) in one litre of media.
5. Carefully pour the mixture into sterile petri dishes by lifting the lid enough only to pour in the agar to avoid contamination.
6. Allow it to cool and solidify for 20 min.
7. Surface-disinfect the seed by pretreating for 1 min in a 1% sodium hypochlorite (NaOCl) solution prepared by diluting 20 parts of laundry bleach (5.25% NaOCl) with 85 parts of water.
8. Place about 10 seeds (depending on size) on the agar surface with a forceps.
9. Incubate the petri dishes at 20–25°C for about 10 days.
10. Identify the seedborne pathogens on the basis of colony and spore characteristics
Near ultraviolet light with a wavelength 300-380 nm (also called black light) may be required to promote sporulation.
The Bacterial Testing Protocol consist of a seed wash assay presented below:
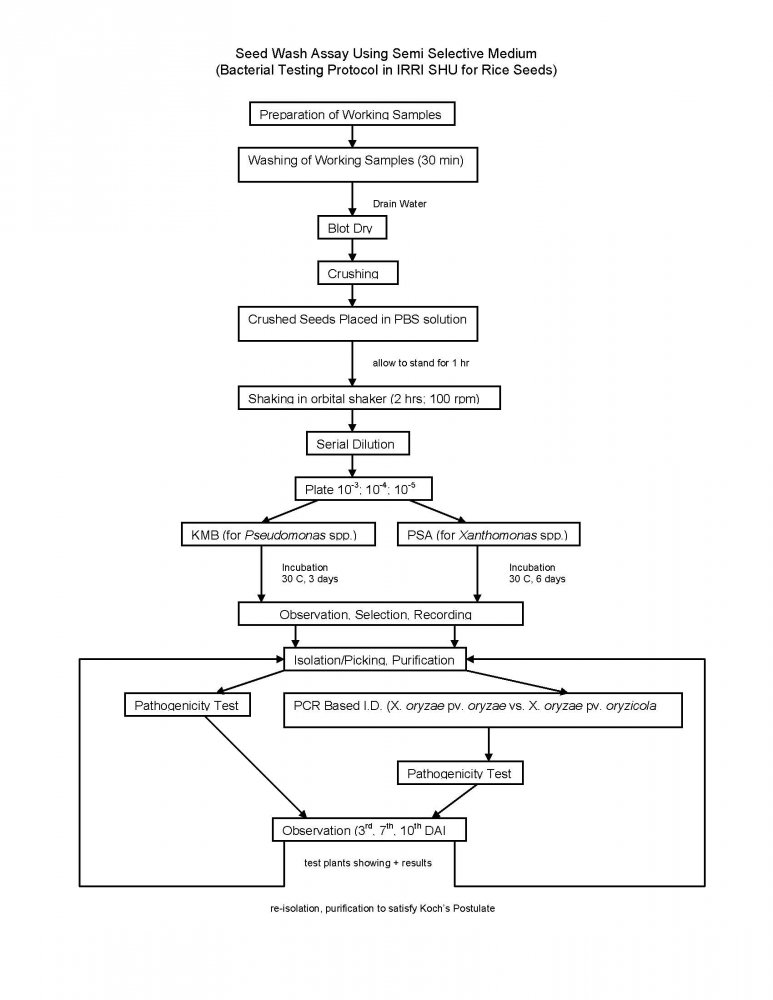
General seed treatment in place at SHU include: Seed fumigation, hot water treatment of rice seed, slurry treatment with benomyl and mancozeb, sodium hypochlorite seed washing and crop health monitoring.
References and further reading
Mew TW, Misra JK. 1994. A manual of Rice Seed Health Testing. IRRI, Manila, The Philippines, 114,pp.
Mew TW, Gonzales P. 2002. A handbook of Rice Seedborne Fungi. Los Baños (Philippines): IRRI, and Enfield N.H.(USA) Science Publishers Inc. 83 pp. [online] Available from URL:http://www.knowledgebank.irri.org Date accessed 07 April 2010
Comments
- No comments found





Leave your comments
Post comment as a guest